Acute pyogenic vertebral osteomyelitis is often due to hematogenous spread of aerobic bacteria.1-4 Conversely, only 0.5% of anaerobic bacteremias lead to osteomyelitis.5 Anaerobic osteomyelitis typically results from the contiguous spread of polymicrobial infections through breaks in the gut mucosal barrier and involves the vertebral bodies in only 2% to 5% of cases.5,6 Although Bacteroides fragilis (B fragilis) is the most common anaerobic pathogen cultivated from blood, accounting for about half of all anaerobic blood isolates, it seldom leads to osteomyelitis.1,2,7-11 We report an uncommon case of B fragilis bacteremia and vertebral osteomyelitis confounded by uncertainties in anaerobic identification and susceptibilities.
Case Presentation
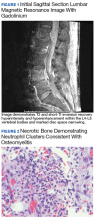
A healthy-appearing male aged 55 years presented to the Naval Medical Center Portsmouth (NMCP) with subacute low back pain and fevers of 103 °F for > 3 weeks. While traveling 4 weeks prior, he completed a course of oseltamivir for influenza B infection; afterward, he was diagnosed with community-acquired pneumonia and treated with a dose of ceftriaxone and a 7-day course of doxycycline. The patient presented to the same facility a week later for low back pain and nonresolving respiratory symptoms, and his therapy was changed to azithromycin, cefuroxime, prednisone, and inhalers. Additionally, after being treated for influenza, he developed constipation and hematochezia for which he did not seek care. The hematochezia was similar to a previous episode from an anal fissure 1 year prior that resolved with stool softeners. When he was finally seen at NMCP after 3 weeks of worsening back pain and fevers, lumbosacral magnetic resonance imaging (MRI) demonstrated vertebral osteomyelitis and discitis at L4-L5 and admitted to the hospital (Figure 1).
After a fluoroscopy-guided biopsy of the L4 vertebral body on hospital day 1, the patient was started on cefepime and vancomycin. The biopsy sample was inoculated onto solid media (blood agar, chocolate agar, and MacConkey agar) and incubated at 36 °C for 24 hours in a 5% CO2 atmosphere, as well as onto Shaedler agar with vitamin K and chopped meat glucose broth and incubated at 36 °C for 48 hours under anaerobic conditions. Metronidazole was added and vancomycin discontinued after 2 anaerobic blood culture vials obtained on hospital day 1, incubated in a Becton Dickinson BACTEC FX automated system, which demonstrated Gram-negative bacilli after 48 hours. The blood culture isolates demonstrated a > 99% probability of being identified as ß-lactamase positive Prevotella loescheii using Thermo Fischer Scientific RapID ANA II biochemical testing. Nitrocefinase discs were used to detect ß-lactamase activity.
The biopsy demonstrated nongranulomatous focal areas of necrotic bone and neutrophilia in a hematopoietic background consistent with acute osteomyelitis (Figure 2); on hospital day 4, ß-lactamase positive B fragilis grew from the bone culture. Additionally, 1 anaerobic vial from a surveillance blood culture set that was obtained on hospital day 3 grew ß-lactamasepositive B fragilis using the same identification methods. With these results he was thought to have a polymicrobial infection (B fragilis and Prevotella loescheii [P loescheii]) from a suspected bowel source based on his hematochezia and history of anal fissure. No aerobic, Gram-negative enterobacteriaceae were isolated, but he had previously been on cefuroxime, which has potential activity against these organisms, for ≥ 2 weeks prior to hospitalization and cultures. He was discharged on moxifloxacin and metronidazole pending final culture results, including requested anaerobic susceptibility testing.
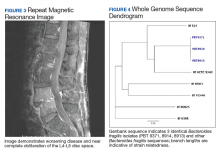
At 1-week follow-up, both aerobic and anaerobic vials from surveillance blood cultures remained negative for any microbes, so antibiotics were deescalated to moxifloxacin monotherapy. However, after 3 days the patient was readmitted for increasing C-reactive protein (CRP) levels and intractable back pain with worsening bilateral radiculopathy. A repeat MRI demonstrated interval disease progression with near obliteration of the L4-L5 disc space and hyperenhancement of the prevertebral soft tissues and adjacent psoas musculature without focal rim-enhancing fluid collection (Figure 3). After repeat L4 biopsy, metronidazole was restarted and ertapenem added for enterobacteriaceae coverage, given the known B fragilis and potential suppression from previous cephalosporin therapy; moxifloxacin was discontinued. L4 biopsy cultures showed no growth, and CRP levels trended down from 154.2 mg/L (start of first admission) to 42.4 mg/L (start of second admission) to 14.9 mg/L (day of discharge) (reference range, 5-9.9 mg/L). He was discharged on ertapenem and metronidazole. He completed a 6-week course without further complication.
During antibiotic therapy he had an unremarkable colonoscopy, CRP normalized to 2.6 mg/L (reference range, 0-4.9 mg/L), and he underwent successful L4-L5 transforaminal lumbar interbody fusion 2 weeks after finishing antibiotics.
We retroactively sent both P loescheii isolates and the 1 B fragilis isolate that grew from the surveillance blood culture to the Multidrug-resistant Organism Repository and Surveillance Network (MRSN) at the Walter Reed Army Institute of Research for identification confirmation and susceptibility analysis. Whole genome sequencing with single nucleotide polymorphism (SNP)-based analysis revealed all isolates were 100% identical and consistent with B fragilis and not P loescheii, based on clustering around other B fragilis sequences found in the National Center for Biotechnology Information (NCBI) Genbank database (Figure 4). All isolates carried the antibiotic resistance genes— cepA, sul(2), tetQ— encoding for possible resistance to cephalosporins, sulphonamides, and tetracyclines, respectively; as well as a point mutation in the gyrA gene (Ser82Phe). None of the isolates carried the nim gene, and screening for the 3 subtypes of B fragilis enterotoxin gene (bft-1, bft-2, bft-3) was negative. Eventual susceptibility testing at the Mayo Clinic several months after the conclusion of the case indicated that the B fragilis isolate was sensitive to piperacillin-tazobactam, ertapenem, clindamycin, and metronidazole; however, testing was not performed against moxifloxacin.