Cross-contamination of pathology specimens is a rare but nonnegligible source of potential morbidity in clinical practice. Contaminant tissue fragments, colloquially referred to as floaters, typically are readily identifiable based on obvious cytomorphologic differences, especially if the tissues arise from different organs; however, one cannot rely on such distinctions in a pathology laboratory dedicated to a single organ system (eg, dermatopathology). The inability to identify quickly and confidently the presence of a contaminant puts the patient at risk for misdiagnosis, which can lead to unnecessary morbidity or even mortality in the case of cancer misdiagnosis. Studies that have been conducted to estimate the incidence of this type of error have suggested an overall incidence rate between approximately 1% and 3%.1,2 Awareness of this phenomenon and careful scrutiny when the histopathologic evidence diverges considerably from the clinical impression is critical for minimizing the negative outcomes that could result from the presence of contaminant tissue. We present a case in which cross-contamination of a pathology specimen led to an initial erroneous diagnosis of an aggressive cutaneous melanoma in a patient with a benign adnexal neoplasm.
Case Report
A 72-year-old man was referred to the Pigmented Lesion and Melanoma Program at Stanford University Medical Center and Cancer Institute (Palo Alto, California) for evaluation and treatment of a presumed stage IIB melanoma on the right preauricular cheek based on a shave biopsy that had been performed (<1 month prior) by his local dermatology provider and subsequently read by an affiliated out-of-state dermatopathology laboratory. Per the clinical history that was gathered at the current presentation, neither the patient nor his wife had noticed the lesion prior to his dermatology provider pointing it out on the day of the biopsy. Additionally, he denied associated pain, bleeding, or ulceration. According to outside medical records, the referring dermatology provider described the lesion as a 4-mm pink pearly papule with telangiectasia favoring a diagnosis of basal cell carcinoma, and a diagnostic shave biopsy was performed. On presentation to our clinic, physical examination of the right preauricular cheek revealed a 4×3-mm depressed erythematous scar with no evidence of residual pigmentation or nodularity (Figure 1). There was no clinically appreciable regional lymphadenopathy.
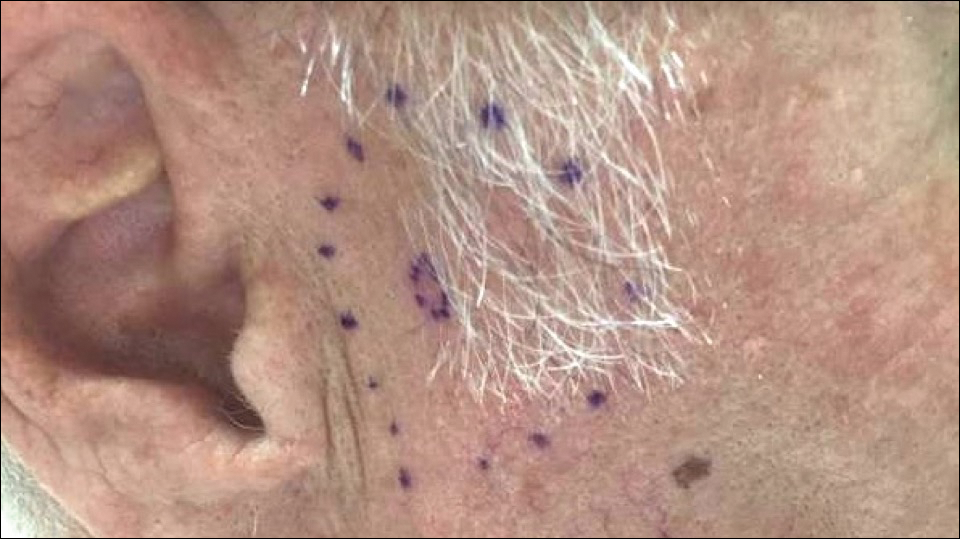
Figure 1. On physical examination at our clinic, a small pink scar (inner broken line) from a prior shave biopsy was noted on the patient’s right cheek. The outer broken line represents the proposed margins for wide local excision based on the initial diagnosis of a clinical stage IIB cutaneous melanoma.
The original dermatopathology report indicated an invasive melanoma with the following pathologic characteristics: superficial spreading type, Breslow depth of at least 2.16 mm, ulceration, and a mitotic index of 8 mitotic figures/mm2 with transection of the invasive component at the peripheral and deep margins. There was no evidence of regression, perineural invasion, lymphovascular invasion, or microsatellites. Interestingly, the report indicated that there also was a basaloid proliferation with features of cylindroma in the same pathology slide adjacent to the aggressive invasive melanoma that was described. Given the complexity of cases referred to our academic center, the standard of care includes internal dermatopathology review of all outside pathology specimens. This review proved critical to this patient’s care in light of the considerable divergence of the initial pathologic diagnosis and the reported clinical features of the lesion.
Internal review of the single pathology slide received from the referring provider showed a total of 4 sections, 3 of which are shown here (Figure 2A). Three sections, including the one not shown, were all consistent with a diagnosis of cylindroma and showed no evidence of a melanocytic proliferation (Figure 2B). However, the fourth section demonstrated marked morphologic dissimilarity compared to the other 3 sections. This outlier section showed a thick cutaneous melanoma with a Breslow depth of at least 2.1 mm, ulceration, a mitotic rate of 12 mitotic figures/mm2, and broad transection of the invasive component at the peripheral and deep margins (Figures 2C and 2D). Correlation with the gross description of tissue processing on the original pathology report indicating that the specimen had been trisected raised suspicion that the fourth and very dissimilar section could be a contaminant from another source that was incorporated into our patient’s histologic sections during processing. Taken together, these discrepancies made the diagnosis of cylindroma alone far more likely than cutaneous melanoma, but we needed conclusive evidence given the dramatic difference in prognosis and management between a cylindroma and an aggressive cutaneous melanoma.
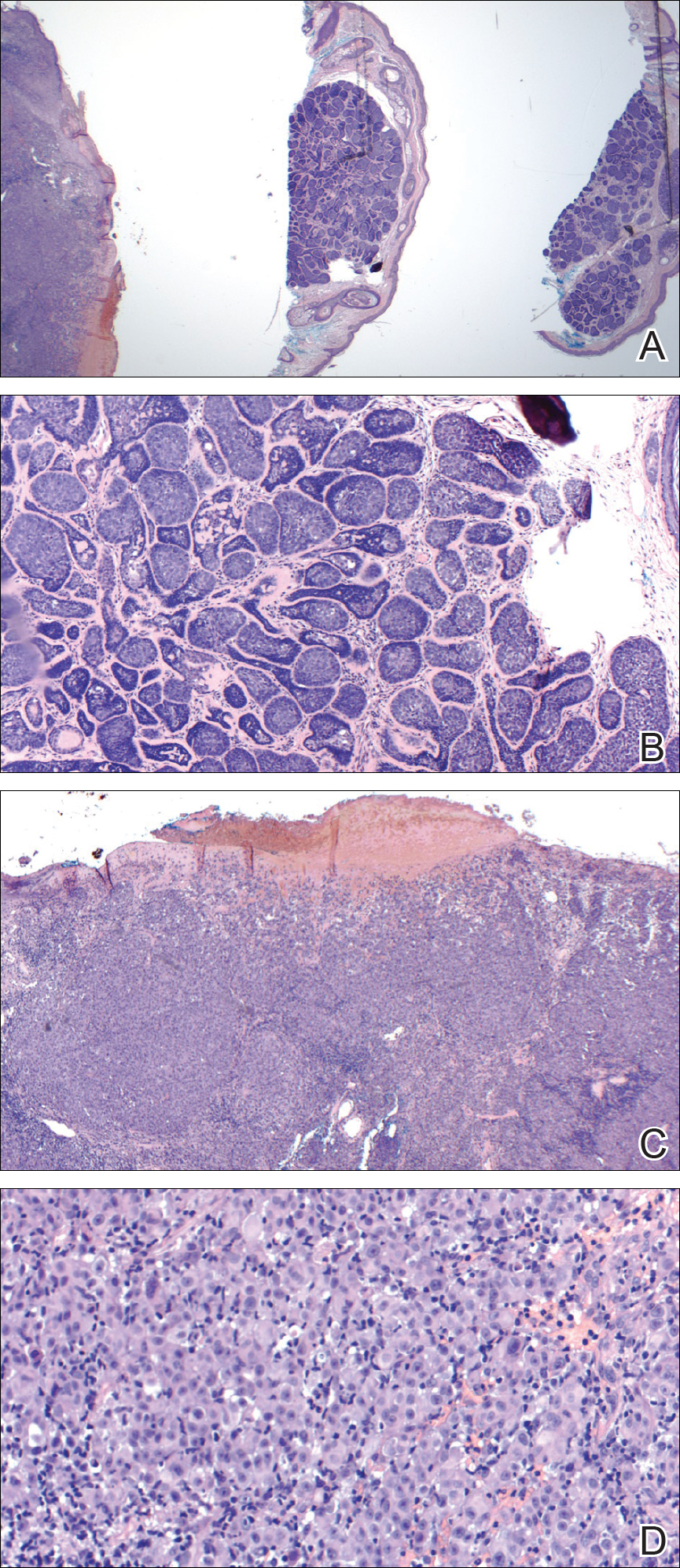
Figure 2. Upon review of 3 of 4 total sections on a single slide received from the dermatopathology laboratory where the specimen was processed, a malignant melanocytic neoplasm with epidermal ulceration was revealed (left), while 3 sections (middle and right as well as one not pictured due to image constraints) showed a benign basaloid neoplasm without epidermal ulceration (A)(H&E, original magnification ×2). On higher power, the middle section demonstrated a basaloid proliferation of well-differentiated cells in the dermis, which supported a diagnosis of cylindroma (B)(H&E, original magnification ×4), and the left section demonstrated a malignant melanocytic proliferation consisting of nested pleomorphic cells without maturation, which supported the diagnosis of invasive melanoma with ulceration (C)(H&E, original magnification ×4). Note the nested and pleomorphic characteristics of the densely packed melanocytes in the invasive melanoma (D)(H&E, original magnification ×20).
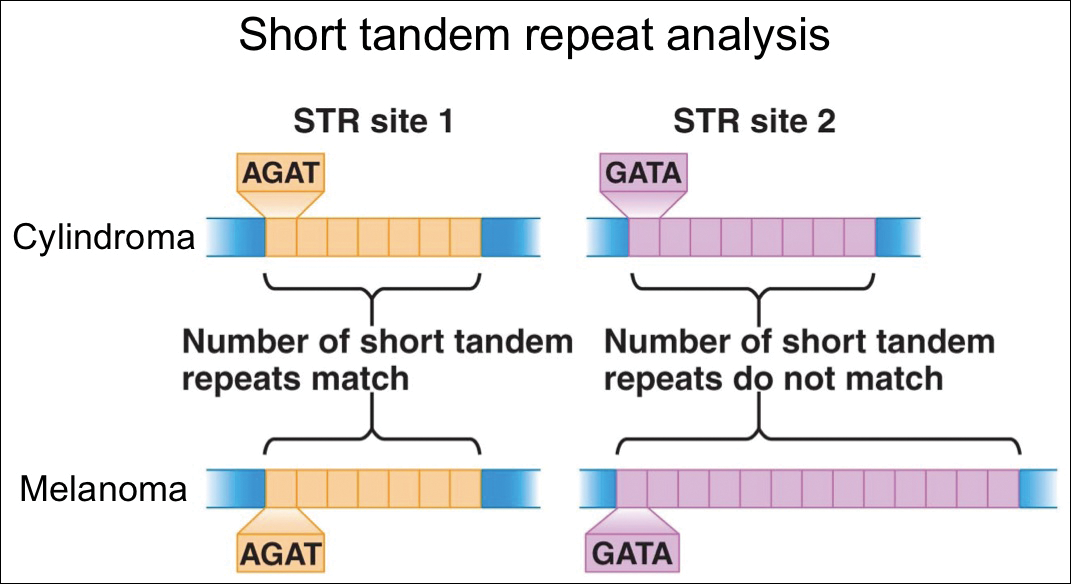
For further diagnostic clarification, we performed polymorphic short tandem repeat (STR) analysis, a well-described forensic pathology technique, to determine if the melanoma and cylindroma specimens derived from different patients, as we hypothesized. This analysis revealed differences in all but one DNA locus tested between the cylindroma specimen and the melanoma specimen, confirming our hypothesis (Figure 3). Subsequent discussion of the case with staff from the dermatopathology laboratory that processed this specimen provided further support for our suspicion that the invasive melanoma specimen was part of a case processed prior to our patient’s benign lesion. Therefore, the wide local excision for treatment of the suspected melanoma fortunately was canceled, and the patient did not require further treatment of the benign cylindroma. The patient expressed relief and gratitude for this critical clarification and change in management.